Nano – rheology of enzymes
Giovanni Zocchi
Dept. of Physics and Astronomy, UCLA
Enzymes couple a chemical process to conformational motion. While end states are often known structurally, a dynamic description of conformational motion is almost entirely lacking. However, it is in the dynamics that some universality may emerge.
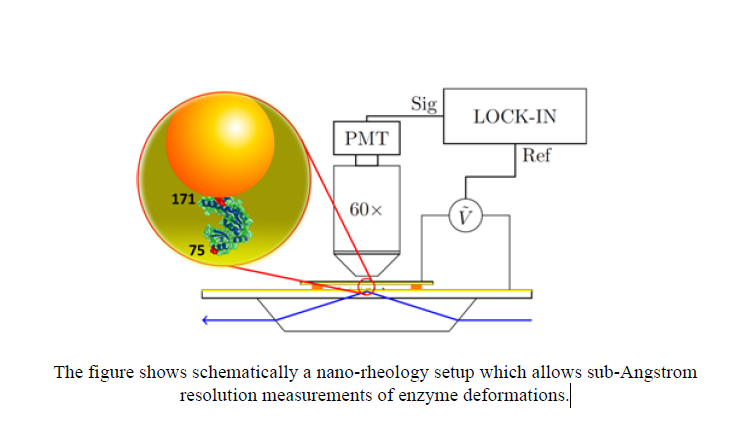
We have developed a nanorheology method where the ensemble averaged deformation of an enzyme subjected to an oscillatory stress is measured with subAngstrom resolution – an improvement of a factor 100 over previous mechanical measurements, giving access to the rheology of the folded state. Measurements on the enzyme Guanylate Kinase reveal a viscoelastic transition in the dynamics. We propose that ligand induced conformational changes generally operate in this viscoelastic regime: the enzyme “flows” from one solid – like conformation to another. It appears that the molecules we are made of behave dynamically like “silly putty”!
View PDF


